By agreement with the publisher, this book is accessible by the search feature, but cannot be browsed.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002.

Molecular Biology of the Cell. 4th edition.
Show detailsIn the previous section, we described the basic components of genetic switches—gene regulatory proteins and the specific DNA sequences that these proteins recognize. We shall now discuss how these components operate to turn genes on and off in response to a variety of signals.
Only 40 years ago the idea that genes could be switched on and off was revolutionary. This concept was a major advance, and it came originally from the study of how E. coli bacteria adapt to changes in the composition of their growth medium. Parallel studies on the lambda bacteriophage led to many of the same conclusions and helped to establish the underlying mechanism. Many of the same principles apply to eucaryotic cells. However, the enormous complexity of gene regulation in higher organisms, combined with the packaging of their DNA into chromatin, creates special challenges and some novel opportunities for control—as we shall see. We begin with the simplest example—an on-off switch in bacteria that responds to a single signal.
The Tryptophan Repressor Is a Simple Switch That Turns Genes On and Off in Bacteria
The chromosome of the bacterium E. coli, a single-celled organism, consists of a single circular DNA molecule of about 4.6 × 106 nucleotide pairs. This DNA encodes approximately 4300 proteins, although only a fraction of these are made at any one time. The expression of many of them is regulated according to the available food in the environment. This is illustrated by the five E. coli genes that code for enzymes that manufacture the amino acid tryptophan. These genes are arranged as a single operon; that is, they are adjacent to one another on the chromosome and are transcribed from a single promoter as one long mRNA molecule (Figure 7-33). But when tryptophan is present in the growth medium and enters the cell (when the bacterium is in the gut of a mammal that has just eaten a meal of protein, for example), the cell no longer needs these enzymes and shuts off their production.

Figure 7-33
The clustered genes in E. coli that code for enzymes that manufacture the amino acid tryptophan. These five genes are transcribed as a single mRNA molecule, a feature that allows their expression to be controlled coordinately. Clusters of genes transcribed (more...)
The molecular basis for this switch is understood in considerable detail. As described in Chapter 6, a promoter is a specific DNA sequence that directs RNA polymerase to bind to DNA, to open the DNA double helix, and to begin synthesizing an RNA molecule. Within the promoter that directs transcription of the tryptophan biosynthetic genes lies a regulating element called an operator (see Figure 7-33). This is simply a short region of regulatory DNA of defined nucleotide sequence that is recognized by a repressor protein, in this case the tryptophan repressor, a member of the helix-turn-helix family (see Figure 7-14). The promoter and operator are arranged so that when the tryptophan repressor occupies the operator, it blocks access to the promoter by RNA polymerase, thereby preventing expression of the tryptophan-producing enzymes (Figure 7-34).

Figure 7-34
Switching the tryptophan genes on and off. If the level of tryptophan inside the cell is low, RNA polymerase binds to the promoter and transcribes the five genes of the tryptophan (trp) operon. If the level of tryptophan is high, however, the tryptophan (more...)
The block to gene expression is regulated in an ingenious way: to bind to its operator DNA, the repressor protein has to have two molecules of the amino acid tryptophan bound to it. As shown in Figure 7-35, tryptophan binding tilts the helix-turn-helix motif of the repressor so that it is presented properly to the DNA major groove; without tryptophan, the motif swings inward and the protein is unable to bind to the operator. Thus the tryptophan repressor and operator form a simple device that switches production of the tryptophan biosynthetic enzymes on and off according to the availability of free tryptophan. Because the active, DNA-binding form of the protein serves to turn genes off, this mode of gene regulation is called negative control, and the gene regulatory proteins that function in this way are called transcriptional repressors or gene repressor proteins.

Figure 7-35
The binding of tryptophan to the tryptophan repressor protein changes the conformation of the repressor. The conformational change enables this gene regulatory protein to bind tightly to a specific DNA sequence (the operator), thereby blocking transcription (more...)
Transcriptional Activators Turn Genes On
We saw in Chapter 6 that purified E. coli RNA polymerase (including the σ subunit) can bind to a promoter and initiate DNA transcription. Some bacterial promoters, however, are only marginally functional on their own, either because they are recognized poorly by RNA polymerase or because the polymerase has difficulty opening the DNA helix and beginning transcription. In either case these poorly functioning promoters can be rescued by gene regulatory proteins that bind to a nearby site on the DNA and contact the RNA polymerase in a way that dramatically increases the probability that a transcript will be initiated. Because the active, DNA-binding form of such a protein turns genes on, this mode of gene regulation is called positive control, and the gene regulatory proteins that function in this manner are known as transcriptional activators or gene activator proteins. In some cases, bacterial gene activator proteins aid RNA polymerase in binding to the promoter by providing an additional contact surface for the polymerase. In other cases, they facilitate the transition from the initial DNA-bound conformation of polymerase to the actively transcribing form, perhaps by stabilizing a transition state.
As in negative control by a transcriptional repressor, a transcriptional activator can operate as part of a simple on-off genetic switch. The bacterial activator protein CAP (catabolite activator protein), for example, activates genes that enable E. coli to use alternative carbon sources when glucose, its preferred carbon source, is not available. Falling levels of glucose induce an increase in the intracellular signaling molecule cyclic AMP, which binds to the CAP protein, enabling it to bind to its specific DNA sequence near target promoters and thereby turn on the appropriate genes. In this way the expression of a target gene is switched on or off, depending on whether cyclic AMP levels in the cell are high or low, respectively. Figure 7-36 summarizes the different ways that positive and negative control can be used to regulate genes.
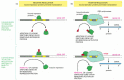
Figure 7-36
Summary of the mechanisms by which specific gene regulatory proteins control gene transcription in procaryotes. (A) Negative regulation; (B) positive regulation. Note that the addition of an “inducing” ligand can turn on a gene either (more...)
In many respects transcriptional activators and transcriptional repressors are similar in design. The tryptophan repressor and the transcriptional activator CAP, for example, both use a helix-turn-helix motif (see Figure 7-14) and both require a small cofactor in order to bind DNA. In fact, some bacterial proteins (including CAP and the bacteriophage lambda repressor) can act as either activators or repressors, depending on the exact placement of the DNA sequence they recognize in relation to the promoter: if the binding site for the protein overlaps the promoter, the polymerase cannot bind and the protein acts as a repressor (Figure 7-37).

Figure 7-37
Some bacterial gene regulatory proteins can act as both a transcriptional activator and a repressor, depending on the precise placement of its binding sites in DNA. An example is the bacteriophage lambda repressor. For some genes, the protein acts as (more...)
A Transcriptional Activator and a Transcriptional Repressor Control the lac Operon
More complicated types of genetic switches combine positive and negative controls. The lac operon in E. coli, for example, unlike the trp operon, is under both negative and positive transcriptional controls by the lac repressor protein and CAP, respectively. The lac operon codes for proteins required to transport the disaccharide lactose into the cell and to break it down. CAP, as we have seen, enables bacteria to use alternative carbon sources such as lactose in the absence of glucose. It would be wasteful, however, for CAP to induce expression of the lac operon if lactose is not present, and the lac repressor ensures that the lac operon is shut off in the absence of lactose. This arrangement enables the control region of lac operon to respond to and integrate two different signals, so that the operon is highly expressed only when two conditions are met: lactose must be present and glucose must be absent. Any of the other three possible signal combinations maintain the cluster of genes in the off state (Figure 7-38).

Figure 7-38
Dual control of the lac operon. Glucose and lactose levels control the initiation of transcription of the lac operon through their effects on the lac repressor protein and CAP. Lactose addition increases the concentration of allolactose, which binds to (more...)
The simple logic of this genetic switch first attracted the attention of biologists over 50 years ago. As explained above, the molecular basis of the switch was uncovered by a combination of genetics and biochemistry, providing the first insight into how gene expression is controlled. Although the same basic strategies are used to control gene expression in higher organisms, the genetic switches that are used are usually much more complex.
Regulation of Transcription in Eucaryotic Cells Is Complex
The two-signal switching mechanism that regulates the lac operon is elegant and simple. However, it is difficult to imagine how it could grow in complexity to allow dozens of signals to regulate transcription from the operon: there is not enough room in the neighborhood of the promoter to pack in a sufficient number of regulatory DNA sequences. How then have eucaryotes overcome such limitations to create their more complex genetic switches?
The regulation of transcription in eucaryotes differs in three important ways from that typically found in bacteria.
- First, eucaryotes make use of gene regulatory proteins that can act even when they are bound to DNA thousands of nucleotide pairs away from the promoter that they influence, which means that a single promoter can be controlled by an almost unlimited number of regulatory sequences scattered along the DNA.
- Second, as we saw in the last chapter, eucaryotic RNA polymerase II, which transcribes all protein-coding genes, cannot initiate transcription on its own. It requires a set of proteins called general transcription factors, which must be assembled at the promoter before transcription can begin. (The term “general” refers to the fact that these proteins assemble on all promoters transcribed by RNA polymerase II; in this they differ from gene regulatory proteins, which act only at particular genes.) This assembly process provides, in principle, multiple steps at which the rate of transcription initiation can be speeded up or slowed down in response to regulatory signals, and many eucaryotic gene regulatory proteins influence these steps.
Having discussed the general transcription factors for RNA polymerase II in Chapter 6 (see pp. 309–312), we focus here on the first and third of these features and how they are used to control eucaryotic gene expression selectively.
Eucaryotic Gene Regulatory Proteins Control Gene Expression from a Distance
Like bacteria, eucaryotes use gene regulatory proteins (activators and repressors) to regulate the expression of their genes but in a somewhat different way. The DNA sites to which the eucaryotic gene activators bound were originally termed enhancers, since their presence “enhanced,” or increased, the rate of transcription dramatically. It came as a surprise when, in 1979, it was discovered that these activator proteins could be bound thousands of nucleotide pairs away from the promoter. Moreover, eucaryotic activators could influence transcription of a gene when bound either upstream or downstream from it. How do enhancer sequences and the proteins bound to them function over these long distances? How do they communicate with the promoter?
Many models for “action at a distance” have been proposed, but the simplest of these seems to apply in most cases. The DNA between the enhancer and the promoter loops out to allow the activator proteins bound to the enhancer to come into contact with proteins (RNA polymerase, one of the general transcription factors, or other proteins) bound to the promoter (see Figure 6-19). The DNA thus acts as a tether, helping a protein bound to an enhancer even thousands of nucleotide pairs away to interact with the complex of proteins bound to the promoter (Figure 7-39). This phenomenon also occurs in bacteria, although less commonly and over much shorter lengths of DNA (Figure 7-40).

Figure 7-39
Binding of two proteins to separate sites on the DNA double helix can greatly increase their probability of interacting. (A) The tethering of one protein to the other via an intervening DNA loop of 500 nucleotide pairs increases their frequency of collision. (more...)

Figure 7-40
Gene activation at a distance. (A) NtrC is a bacterial gene regulatory protein that activates transcription by facilitating the transition between the initial binding of RNA polymerase to the promoter and the formation of an initiating complex (discussed (more...)
A Eucaryotic Gene Control Region Consists of a Promoter Plus Regulatory DNA Sequences
Because eucaryotic gene regulatory proteins can control transcription when bound to DNA far away from the promoter, the DNA sequences that control the expression of a gene are often spread over long stretches of DNA. We shall use the term gene control region to refer to the whole expanse of DNA involved in regulating transcription of a gene, including the promoter, where the general transcription factors and the polymerase assemble, and all of the regulatory sequences to which gene regulatory proteins bind to control the rate of the assembly processes at the promoter (Figure 7-41). In higher eucaryotes it is not unusual to find the regulatory sequences of a gene dotted over distances as great as 50,000 nucleotide pairs. Although much of this DNA serves as “spacer” sequence and is not recognized by gene regulatory proteins, this spacer DNA may facilitate transcription by providing the flexibility needed for communication between DNA-bound proteins. It is also important to keep in mind that, like other regions of eucaryotic chromosomes, much of the DNA in gene control regions is packaged into nucleosomes and higher-order forms of chromatin, thereby compacting its length.
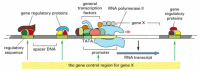
Figure 7-41
The gene control region of a typical eucaryotic gene. The promoter is the DNA sequence where the general transcription factors and the polymerase assemble (see Figure 6-16). The regulatory sequences serve as binding sites for gene regulatory proteins, (more...)
In this chapter we generally use the term gene to refer only to a segment of DNA that is transcribed into RNA (see Figure 7-41). However, the classical view of a gene would include the gene control region as well. The different definitions arise from the different ways in which genes were historically identified. The discovery of alternative RNA splicing has further complicated the definition of a gene—a point we discussed briefly in Chapter 6 and will return to later in this chapter.
Although many gene regulatory proteins bind to enhancer sequences and activate gene transcription, many others function as negative regulators, as we see below. In contrast to the small number of general transcription factors, which are abundant proteins that assemble on the promoters of all genes transcribed by RNA polymerase II, there are thousands of different gene regulatory proteins. For example, of the roughly 30,000 human genes, an estimated 5–10% encode gene regulatory proteins. These regulatory proteins vary from one gene control region to the next, and each is usually present in very small amounts in a cell, often less than 0.01% of the total protein. Most of them recognize their specific DNA sequences using one of the DNA-binding motifs discussed previously, although as we discuss below, some do not recognize DNA directly but instead assemble on other DNA-bound proteins.
The gene regulatory proteins allow the individual genes of an organism to be turned on or off specifically. Different selections of gene regulatory proteins are present in different cell types and thereby direct the patterns of gene expression that give each cell type its unique characteristics. Each gene in a eucaryotic cell is regulated differently from nearly every other gene. Given the number of genes in eucaryotes and the complexity of their regulation, it has been difficult to formulate simple rules for gene regulation that apply in every case. We can, however, make some generalizations about how gene regulatory proteins, once bound to a gene control region on DNA, influence the rate of transcription initiation, as we now explain.
Eucaryotic Gene Activator Proteins Promote the Assembly of RNA Polymerase and the General Transcription Factors at the Startpoint of Transcription
Most gene regulatory proteins that activate gene transcription—that is, most gene activator proteins—have a modular design consisting of at least two distinct domains. One domain usually contains one of the structural motifs discussed previously that recognizes a specific regulatory DNA sequence. In the simplest cases, a second domain—sometimes called an activation domain—accelerates the rate of transcription initiation. This type of modular design was first revealed by experiments in which genetic engineering techniques were used to create a hybrid protein containing the activation domain of one protein fused to the DNA-binding domain of a different protein (Figure 7-42).

Figure 7-42
The modular structure of a gene activator protein. Outline of an experiment that reveals the presence of independent DNA-binding and transcription-activating domains in the yeast gene activator protein Gal4. A functional activator can be reconstituted (more...)
Once bound to DNA, how do eucaryotic gene activator proteins increase the rate of transcription initiation? As we will see shortly, there are several mechanisms by which this can occur, and, in many cases, these different mechanisms work in concert at a single promoter. But, regardless of the precise biochemical pathway, the main function of activators is to attract, position, and modify the general transcription factors and RNA polymerase II at the promoter so that transcription can begin. They do this both by acting directly on the transcription machinery itself and by changing the chromatin structure around the promoter.
We consider first the ways in which activators directly influence the positioning of the general transcription factors and RNA polymerase at promoters and help kick them into action. Although the general transcription factors and RNA polymerase II assemble in a stepwise, prescribed order in vitro (see Figure 6-16), there are cases in living cells where some of them are brought to the promoter as a large pre-assembled complex that is sometimes called the RNA polymerase II holoenzyme. In addition to some of the general transcription factors and RNA polymerase, the holoenzyme typically contains a 20-subunit protein complex called the mediator, which was first identified biochemically as being required for activators to stimulate transcription initiation.
Many activator proteins interact with the holoenzyme complex and thereby make it more energetically favorable for it to assemble on a promoter that is linked through DNA to the site where the activator protein is bound (Figure 7-43A). In this sense, eucaryotic activators resemble those of bacteria in helping to attract and position RNA polymerase on specific sites on DNA (see Figure 7-36). One type of experiment that supports the idea that activators attract the holoenzyme complex to promoters creates an “activator bypass” (Figure 7-43B). Here, a sequence-specific DNA-binding domain is experimentally fused directly to a component of the mediator; this hybrid protein, which lacks an activation domain, strongly stimulates transcription initiation when the DNA sequence to which it binds is placed in proximity to a promoter.

Figure 7-43
Activation of transcription initiation in eucaryotes by recruitment of the eucaryotic RNA polymerase II holoenzyme complex. (A) An activator protein bound in proximity to a promoter attracts the holoenzyme complex to the promoter. According to this model, (more...)
Although recruitment of the holoenzyme complex to promoters provides a conceptually simple mechanism for envisioning gene activation, the effect of activators on the holoenzyme complex is probably more complicated. For example, a stepwise assembly of the general transcription factors (see Figure 6-16) may occur on some promoters. On others, their rearrangement, once brought to DNA as part of the holoenzyme, may be required. In addition, most forms of the holoenzyme complex lacks some of the general transcription factors (notably TFIID and TFIIA), and these must be assembled on the promoter separately (see Figure 7-43A). In principle, any of these assembly processes could be a slow step on the pathway to transcription initiation, and activators could facilitate their completion. In fact, many activators have been shown to interact with one or more of the general transcription factors, and several have been shown to directly accelerate their assembly at the promoter (Figure 7-44).
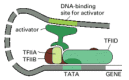
Figure 7-44
A model for the action of some eucaryotic transcriptional activators. The gene activator protein, bound to DNA in the rough vicinity of the promoter, facilitates the assembly of some of the general transcription factors. Although some activator proteins (more...)
Eucaryotic Gene Activator Proteins Modify Local Chromatin Structure
In addition to their direct actions in assembling the RNA polymerase holoenzyme and the general transcription factors on DNA, gene activator proteins also promote transcription initiation by changing the chromatin structure of the regulatory sequences and promoters of genes. As we saw in Chapter 4, the two most important ways of locally altering chromatin structure are through covalent histone modifications and nucleosome remodeling (see Figures 4-34 and 4-35). Many gene activator proteins make use of both these mechanisms by binding to and thereby recruiting histone acetyl transferases (HATs), commonly known as histone acetylases, and ATP-dependent chromatin remodeling complexes (Figure 7-45) to work on nearby chromatin. In general terms, the local alterations in chromatin structure that ensue allow greater accessibility to the underlying DNA. This accessibility facilitates the assembly of the general transcription factors and the RNA polymerase holoenzyme at the promoter, and it also allows the binding of additional gene regulatory proteins to the control region of the gene (Figure 7-46A).

Figure 7-45
Local alterations in chromatin structure directed by eucaryotic gene activator proteins. Histone acetylation and nucleosome remodeling generally render the DNA packaged in chromatin more accessible to other proteins in the cell, including those required (more...)
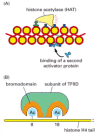
Figure 7-46
Two specific ways that local histone acetylation can stimulate transcription initiation. (A) Some gene activator proteins can bind directly to DNA that is packaged in unmodified chromatin. By attracting histone acetylases (and nucleosome remodeling complexes), (more...)
The general transcription factors seem unable to assemble onto a promoter that is packaged in a conventional nucleosome. In fact, such packaging may have evolved in part to ensure that leaky, or basal, transcription initiation (initiation at a promoter in the absence of gene activator protein bound upstream of it) does not occur. As well as making the DNA more generally accessible, local histone acetylation has a more specialized role in promoting transcription initiation. As discussed in Chapter 4 (see Figure 4-35), certain patterns of histone acetylation are associated with transcriptionally active chromatin, and gene activator proteins, by recruiting histone acetylases, produce these patterns. One such pattern (Figure 7-46B) is directly recognized by one of the subunits of the general transcription factor TFIID, and this recognition apparently helps the factor assemble DNA that is packaged in chromatin. Thus gene activator proteins, through the action of histone acetylases, can indirectly aid in the assembly of the general transcription factors at a promoter and thereby stimulate transcription initiation.
Gene Activator Proteins Work Synergistically
We have seen that eucaryotic gene activator proteins can influence several different steps in transcription initiation, and this property has important consequences when different activator proteins work together. In general, where several factors work together to enhance a reaction rate, the joint effect is generally not merely the sum of the enhancements caused by each factor alone, but the product. If, for example, factor A lowers the free-energy barrier for a reaction by a certain amount and thereby speeds up the reaction 100-fold, and factor B, by acting on another aspect of the reaction, does likewise, then A and B acting in parallel will lower the barrier by a double amount and speed up the reaction 10,000-fold. Similar multiplicative effects occur if A and B speed the reaction by each helping to recruit necessary proteins to the reaction site. Thus, gene activator proteins often exhibit what is called transcriptional synergy, where the transcription rate produced by several activator proteins working together is much higher than that produced by any of the activators working alone (Figure 7-47). Transcriptional synergy is observed both between different gene activator proteins bound upstream of a gene and between multiple DNA-bound molecules of the same activator. It is therefore not difficult to see how multiple gene regulatory proteins, each binding to a different regulatory DNA sequence, could control the final rate of transcription of a eucaryotic gene.

Figure 7-47
Transcriptional synergy. In this experiment, the rate of transcription produced by three experimentally constructed regulatory regions is compared in a eucaryotic cell. Transcriptional synergy, the greater than additive effect of the activators, is observed (more...)
Since gene activator proteins can influence many different steps on the pathway to transcriptional activation, it is worth considering whether these steps always occur in a prescribed order. For example does chromatin remodeling necessarily precede histone acetylation or vice versa? When does recruitment of the holoenzyme complex occur relative to the chromatin modifying steps? The answers to these questions appears to be different for different genes—and even for the same gene under different conditions (Figure 7-48). Whatever the precise mechanisms and the order in which they are carried out, a gene regulatory protein must be bound to DNA either directly or indirectly to influence transcription of its target promoter, and the rate of transcription of a gene ultimately depends upon the spectrum of regulatory proteins bound upstream and downstream of its transcription start site.

Figure 7-48
An order of events leading to transcription initiation at a specific promoter. The well-studied example shown is from a promoter in the budding yeast S. cerevisiae. The chromatin remodeling complex and histone acetylase apparently dissociate from the (more...)
Eucaryotic Gene Repressor Proteins Can Inhibit Transcription in Various Ways
Like bacteria, eucaryotes use gene repressor proteins in addition to activator proteins to regulate transcription of their genes. However, because of differences in the way transcription is initiated in eucaryotes and bacteria, eucaryotic repressors have many more possible mechanisms of action. For example, we saw in Chapter 4 that whole regions of eucaryotic chromosomes can be packaged into heterochromatin, a form of chromatin that is normally resistant to transcription. We will return to this feature of eucaryotic chromosomes later in this chapter. In addition to molecules that shut down large regions of chromatin, eucaryotic cells also contain gene regulatory proteins that act only locally to repress transcription of nearby genes. Unlike bacterial repressors, most do not directly compete with the RNA polymerase for access to the DNA; rather they work by a variety of other mechanisms, some of which are illustrated in Figure 7-49. Like gene activator proteins, many eucaryotic repressor proteins act through more than one mechanism, thereby ensuring robust and efficient repression.

Figure 7-49
Five ways in which eucaryotic gene repressor proteins can operate. (A) Gene activator proteins and gene repressor proteins compete for binding to the same regulatory DNA sequence. (B) Both proteins can bind DNA, but the repressor binds to the activation (more...)
Eucaryotic Gene Regulatory Proteins Often Assemble into Complexes on DNA
So far we have been discussing eucaryotic gene regulatory proteins as though they work as individual polypeptides. In reality, most act as parts of complexes composed of several (and sometimes many) polypeptides, each with a distinct function. These complexes often assemble only in the presence of the appropriate DNA sequence. In some well-studied cases, for example, two gene regulatory proteins with a weak affinity for each other cooperate to bind to a DNA sequence, neither protein having a sufficient affinity for DNA to efficiently bind to the DNA site on its own. Once bound to DNA, the protein dimer creates a distinct surface that is recognized by a third protein that carries an activator domain that stimulates transcription (Figure 7-50). This example illustrates an important general point: protein-protein interactions that are too weak to cause proteins to assemble in solution can cause the proteins to assemble on DNA; in this way the DNA sequence acts as a “crystallization” site or seed for the assembly of a protein complex.

Figure 7-50
Eucaryotic gene regulatory proteins often assemble into complexes on DNA. Seven gene regulatory proteins are shown in (A). The nature and function of the complex they form depends on the specific DNA sequence that seeds their assembly. In (B), some assembled (more...)
An individual gene regulatory protein can often participate in more than one type of regulatory complex. A protein might function, for example, in one case as part of a complex that activates transcription and in another case as part of a complex that represses transcription (see Figure 7-50). Thus individual eucaryotic gene regulatory proteins are not necessarily dedicated activators or repressors; instead, they function as regulatory units that are used to generate complexes whose function depends on the final assembly of all of the individual components. This final assembly, in turn, depends both on the arrangement of control region DNA sequences and on which gene regulatory proteins are present in the cell.
Gene regulatory proteins that do not themselves bind DNA but assemble on DNA-bound gene regulatory proteins are often termed coactivators or corepressors, depending on their effect on transcription initiation. As shown in Figure 7-50, the same coactivator or corepressor can assemble on different DNA binding proteins. Coactivators and corepressors typically carry out multiple functions: they can interact with chromatin remodeling complexes, histone modifying enzymes, the RNA polymerase holoenzyme, and several of the general transcription factors.
In some cases, the precise DNA sequence to which a regulatory protein directly binds can affect the conformation of this protein and thereby influence its subsequent transcriptional activity. When bound to one type of DNA sequence, for example, a steroid hormone receptor interacts with a corepressor and ultimately turns off transcription. When bound to a slightly different DNA sequence, it assumes a different conformation and interacts with a coactivator, thereby stimulating transcription.
Typically, the assembly of a group of regulatory proteins on DNA is guided by a few relatively short stretches of nucleotide sequence (see Figure 7-50). However, in some cases, a more elaborate protein-DNA structure, termed an enhancesome, is formed (Figure 7-51). A hallmark of enhancesomes is the participation of architectural proteins that bend the DNA by a defined angle and thereby promote the assembly of the other enhancesome proteins. Since formation of the enhancesome requires the presence of many gene regulatory proteins, it provides a simple way to ensure that a gene is expressed only when the correct combination of these proteins is present in the cell. We saw earlier how the formation of gene regulatory heterodimers in solution provides a mechanism for the combinatorial control of gene expression. The assembly of larger complexes of gene regulatory proteins on DNA provides a second important mechanism for combinatorial control, offering far richer opportunities.

Figure 7-51
Schematic depiction of an enhancesome. The protein depicted in yellow is termed an architectural protein since its main role is to bend the DNA to allow the cooperative assembly of the other components. The protein surface of this enhancesome interacts (more...)
Complex Genetic Switches That Regulate Drosophila Development Are Built Up from Smaller Modules
Given that gene regulatory proteins can be positioned at multiple sites along long stretches of DNA, that these proteins can assemble into complexes at each site, and that the complexes can influence the chromatin structure and the recruitment and assembly of the general transcription machinery at the promoter, there would seem to be almost limitless possibilities for the elaboration of control devices to regulate eucaryotic gene transcription.
A particularly striking example of a complex, multicomponent genetic switch is that controlling the transcription of the Drosophila even-skipped (eve) gene, whose expression plays an important part in the development of the Drosophila embryo. If this gene is inactivated by mutation, many parts of the embryo fail to form, and the embryo dies early in development. As discussed in Chapter 21, at the earliest stage of development where eve is expressed, the embryo is a single giant cell containing multiple nuclei in a common cytoplasm. This cytoplasm is not uniform, however: it contains a mixture of gene regulatory proteins that are distributed unevenly along the length of the embryo, thus providing positional information that distinguishes one part of the embryo from another (Figure 7-52). (The way these differences are initially set up is discussed in Chapter 21.) Although the nuclei are initially identical, they rapidly begin to express different genes because they are exposed to different gene regulatory proteins. The nuclei near the anterior end of the developing embryo, for example, are exposed to a set of gene regulatory proteins that is distinct from the set that influences nuclei at the posterior end of the embryo.
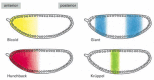
Figure 7-52
The nonuniform distribution of four gene regulatory proteins in an early Drosophila embryo. At this stage the embryo is a syncytium, with multiple nuclei in a common cytoplasm. Although not illustrated in these drawings, all of these proteins are concentrated (more...)
The regulatory DNA sequences of the eve gene are designed to read the concentrations of gene regulatory proteins at each position along the length of the embryo and to interpret this information in such a way that the eve gene is expressed in seven stripes, each initially five to six nuclei wide and positioned precisely along the anterior-posterior axis of the embryo (Figure 7-53). How is this remarkable feat of information processing carried out? Although the molecular details are not yet all understood, several general principles have emerged from studies of eve and other Drosophila genes that are similarly regulated.

Figure 7-53
The seven stripes of the protein encoded by the even-skipped (eve) gene in a developing Drosophila embryo. Two and one-half hours after fertilization, the egg was fixed and stained with antibodies that recognize the Eve protein (green) and antibodies (more...)
The regulatory region of the eve gene is very large (approximately 20,000 nucleotide pairs). It is formed from a series of relatively simple regulatory modules, each of which contains multiple regulatory sequences and is responsible for specifying a particular stripe of eve expression along the embryo. This modular organization of the eve gene control region is revealed by experiments in which a particular regulatory module (say, that specifying stripe 2) is removed from its normal setting upstream of the eve gene, placed in front of a reporter gene (see Figure 7-42), and reintroduced into the Drosophila genome (Figure 7-54A). When developing embryos derived from flies carrying this genetic construct are examined, the reporter gene is found to be expressed in precisely the position of stripe 2 (see Figure 7-54). Similar experiments reveal the existence of other regulatory modules, each of which specifies one of the other six stripes or some part of the expression pattern that the gene displays at later stages of development.

Figure 7-54
Experiment demonstrating the modular construction of the eve gene regulatory region. (A) A 480-nucleotide-pair piece of the eve regulatory region was removed and inserted upstream of a test promoter that directs the synthesis of the enzyme β-galactosidase (more...)
The Drosophila eve Gene Is Regulated by Combinatorial Controls
A detailed study of the stripe 2 regulatory module has provided insights into how it reads and interprets positional information. It contains recognition sequences for two gene regulatory proteins (Bicoid and Hunchback) that activate eve transcription and two (Krüppel and Giant) that repress it (Figure 7-55). (The gene regulatory proteins of Drosophila often have colorful names reflecting the phenotype that results if the gene encoding the protein is inactivated by mutation.) The relative concentrations of these four proteins determine whether protein complexes forming at the stripe 2 module turn on transcription of the eve gene. Figure 7-56 shows the distributions of the four gene regulatory proteins across the region of a Drosophila embryo where stripe 2 forms. Although the precise details are not known, it seems likely that either one of the two repressor proteins, when bound to the DNA, will turn off the stripe 2 module, whereas both Bicoid and Hunchback must bind for its maximal activation. This simple regulatory unit thereby combines these four positional signals so as to turn on the stripe 2 module (and therefore the expression of the eve gene) only in those nuclei that are located where the levels of both Bicoid and Hunchback are high and both Krüppel and Giant are absent. This combination of activators and repressors occurs only in one region of the early embryo; everywhere else, therefore, the stripe 2 module is silent.
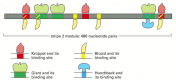
Figure 7-55
Close-up view of the eve stripe 2 unit. The segment of the eve gene control region identified in the previous figure contains regulatory sequences, each of which binds one or another of four gene regulatory proteins. It is known from genetic experiments (more...)

Figure 7-56
Distribution of the gene regulatory proteins responsible for ensuring that eve is expressed in stripe 2. The distributions of these proteins were visualized by staining a developing Drosophila embryo with antibodies directed against each of the four proteins (more...)
We have already discussed two mechanisms of combinatorial control of gene expression—heterodimerization of gene regulatory proteins in solution (see Figure 7-22) and the assembly of combinations of gene regulatory proteins into small complexes on DNA (see Figure 7-50). It is likely that both mechanisms participate in the complex regulation of eve expression. In addition, the regulation of stripe 2 just described illustrates a third type of combinatorial control. Because the individual regulatory sequences in the eve stripe 2 module are strung out along the DNA, many sets of gene regulatory proteins can be bound simultaneously and influence the promoter of a gene. The promoter integrates the transcriptional cues provided by all of the bound proteins (Figure 7-57).

Figure 7-57
Integration at a promoter. Multiple sets of gene regulatory proteins can work together to influence transcription initiation at a promoter, as they do in the eve stripe 2 module illustrated previously in Figure 7-55. It is not yet understood in detail (more...)
The regulation of eve expression is an impressive example of combinatorial control. Seven combinations of gene regulatory proteins—one combination for each stripe—activate eve expression, while many other combinations (all those found in the interstripe regions of the embryo) keep the stripe elements silent. The other stripe regulatory modules are thought to be constructed along lines similar to those described for stripe 2, being designed to read positional information provided by other combinations of gene regulatory proteins. The entire gene control region, strung out over 20,000 nucleotide pairs of DNA, binds more than 20 different proteins. A large and complex control region is thereby built from a series of smaller modules, each of which consists of a unique arrangement of short DNA sequences recognized by specific gene regulatory proteins. Although the details are not yet understood, these gene regulatory proteins are thought to employ a number of the mechanisms previously described for activators and repressors. In this way, a single gene can respond to an enormous number of combinatorial inputs.
Complex Mammalian Gene Control Regions Are Also Constructed from Simple Regulatory Modules
It has been estimated that 5–10% of the coding capacity of a mammalian genome is devoted to the synthesis of proteins that serve as regulators of gene transcription. This large number of genes reflects the exceedingly complex network of controls governing expression of mammalian genes. Each gene is regulated by a set of gene regulatory proteins; each of those proteins is the product of a gene that is in turn regulated by a whole set of other proteins, and so on. Moreover, the regulatory protein molecules are themselves influenced by signals from outside the cell, which can make them active or inactive in a whole variety of ways (Figure 7-58). Thus, pattern of gene expression in a cell can be viewed as the result of a complicated molecular computation that the intracellular gene control network performs in response to information from the cell's surroundings. We shall discuss this further in Chapter 21, dealing with multicellular development, but the complexity is remarkable even at the level of the individual genetic switch, regulating activity of a single gene. It is not unusual, for example, to find a mammalian gene with a control region that is 50,000 nucleotide pairs in length, in which many modules, each containing a number of regulatory sequences that bind gene regulatory proteins, are interspersed with long stretches of spacer DNA.

Figure 7-58
Some ways in which the activity of gene regulatory proteins is regulated in eucaryotic cells. (A) The protein is synthesized only when needed and is rapidly degraded by proteolysis so that it does not accumulate. (B) Activation by ligand binding. (C) (more...)
One of the best-understood examples of a complex mammalian regulatory region is found in the human β-globin gene, which is expressed exclusively in red blood cells and at a specific time in their development. A complex array of gene regulatory proteins controls the expression of the gene, some acting as activators and others as repressors (Figure 7-59). The concentrations (or activities) of many of these gene regulatory proteins are thought to change during development, and only a particular combination of all the proteins triggers transcription of the gene. The human β-globin gene is part of a cluster of globin genes (Figure 7-60A). The five genes of the cluster are transcribed exclusively in erythroid cells, that is, cells of the red blood cell lineage. Moreover, each gene is turned on at a different stage of development (see Figure 7-60B) and in different organs: the ε-globin gene is expressed in the embryonic yolk sac, γ in the yolk sac and the fetal liver, and δ and β primarily in the adult bone marrow. Each of the globin genes has its own set of regulatory proteins that are necessary to turn the gene on at the appropriate time and tissue. In addition to the individual regulation of each of the globin genes, the entire cluster appears to be subject to a shared control region called a locus control region (LCR). The LCR lies far upstream from the gene cluster (see Figure 7-60A), and we shall discuss its function next.
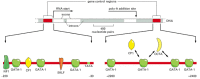
Figure 7-59
Model for the control of the human β-globin gene. The diagram shows some of the gene regulatory proteins thought to control expression of the gene during red blood cell development (see Figure 7-60). Some of the gene regulatory proteins shown, (more...)

Figure 7-60
The cluster of β-like globin genes in humans. (A) The large chromosomal region shown spans 100,000 nucleotide pairs and contains the five globin genes and a locus control region (LCR). (B) Changes in the expression of the β-like globin (more...)
In cells where the globin genes are not expressed (such as brain or skin cells), the whole gene cluster appears tightly packaged into chromatin. In erythroid cells, by contrast, the entire gene cluster is still folded into nucleosomes, but the higher-order packing of the chromatin has become decondensed This change occurs even before the individual globin genes are transcribed, suggesting that there are two steps of regulation. In the first, the chromatin of the entire globin locus becomes decondensed, which is presumed to allow additional gene regulatory proteins access to the DNA. In the second step, the remaining gene regulatory proteins assemble on the DNA and direct the expression of individual genes.
The LCR appears to act by controlling chromatin condensation, and its importance can be seen in patients with a certain type of thalassemia, a severe inherited form of anemia. In these patients, the β-globin locus is found to have undergone deletions that remove all or part of the LCR, and although the β-globin gene and its nearby regulatory regions are intact, the gene remains transcriptionally silent even in erythroid cells. Moreover, the β-globin gene in the erythroid cells fails to undergo the normal chromatin decondensation step that occurs during erythroid cell development.
Many LCRs (that is, DNA regulatory sequences that regulate the accessibility and expression of distant genes or gene clusters) are present in the human genome, and they regulate a wide variety of cell-type specific genes. The way in which they function is not understood in detail, but several models have been proposed. The simplest is based on principles we have already discussed in this chapter: the gene regulatory proteins that bind to the LCR interact through DNA looping with proteins bound to the control regions of the genes they regulate. In this way, the proteins bound at the LCR could attract chromatin remodeling complexes and histone modifying enzymes that could alter the chromatin structure of the locus before transcription begins. Other models for LCRs propose a mechanism by which proteins initially bound at the LCR attract other proteins that assemble cooperatively and therefore spread along the DNA toward the genes they control, modifying the chromatin as they proceed.
Insulators Are DNA Sequences That Prevent Eucaryotic Gene Regulatory Proteins from Influencing Distant Genes
All genes have control regions, which dictate at which times, under what conditions, and in what tissues the gene will be expressed. We also have seen that eucaryotic gene regulatory proteins can act across very long stretches of DNA. How then are control regions of different genes kept from interfering with one another? In other words, what keeps a gene regulatory protein bound on the control region of one gene from inappropriately influencing transcription of adjacent genes?
Several mechanisms have been proposed to account for this regulatory compartmentalization, but the best understood rely on insulator elements, also called boundary elements. Insulator elements (insulators, for short) are DNA sequences that bind specialized proteins and have two specific properties (Figure 7-61). First, they buffer genes from the repressing effects of heterochromatin. When a gene (from a fly or a mouse, for example) and its normal control region is inserted into different positions in the genome, it is often expressed at levels that vary depending on its site of insertion in the genome and are especially low when it is inserted amid heterochromatin. We saw an example of this position effect in Chapter 4, where genes inserted into heterochromatin are transcriptionally silenced (see Figure 4-45). When insulator elements that flank the gene and its control region are included, however, the gene is usually expressed normally, irrespective of its new position in the genome. The second property of insulators is in some sense the converse of this: they can block the action of enhancers (see Figure 7-61). For this to occur, the insulator must be located between the enhancer and the promoter of the target gene.
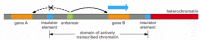
Figure 7-61
Schematic diagram summarizing the properties of insulators. Insulators both prevent the spread of heterochromatin (right-hand side of diagram) and directionally block the action of enhancers (left-hand side). Thus gene B is properly regulated and gene (more...)
Thus insulators can define domains of gene expression, both buffering the gene from outside effects and preventing the control region of the gene (or cluster of genes) from acting outside the domain. For example, the globin LCR (discussed above) is associated with a neighboring insulator which allows the LCR to influence only the cluster of globin genes. Presumably, another insulator is located on the distal side of the globin cluster, serving to define the other end of the domain.
The distribution of insulators in a genome is therefore thought to divide it into independent domains of gene regulation and chromatin structure. Consistent with this idea, the distribution of insulators across a genome is roughly correlated with variations in chromatin structure. For example, an insulator-binding protein from flies is localized preferentially to interbands (and also to the edges of puffs) in polytene chromosomes (Figure 7-62).

Figure 7-62
Localization of a Drosophila insulator-binding protein on polytene chromosomes. A polytene chromosome (see pp. 218–220) was stained with propidium iodide (red) to show its banding patterns—with bands appearing bright red and interbands (more...)
The mechanisms by which insulators work are not currently understood, and different insulators may function in different ways. At least some pairs of insulators may define the basis of a looped chromosomal domain (see Figure 4-44). It has been proposed that chromosomes of all eucaryotes are divided by insulators into independent looped domains, each regulated separately from all the others.
Bacteria Use Interchangeable RNA Polymerase Subunits to Help Regulate Gene Transcription
We have seen the importance of gene regulatory proteins that bind to regulatory sequences in DNA and signal to the transcription apparatus whether or not to start the synthesis of an RNA chain. Although this is the main way of controlling transcriptional initiation in both eucaryotes and procaryotes, some bacteria and their viruses use an additional strategy based on interchangeable subunits of RNA polymerase. As described in Chapter 6, a sigma (σ) subunit is required for the bacterial RNA polymerase to recognize a promoter. Many bacteria make several different sigma subunits, each of which can interact with the RNA polymerase core and direct it to a different set of promoters (Table 7-2). This scheme permits one large set of genes to be turned off and a new set to be turned on simply by replacing one sigma subunit with another; the strategy is efficient because it bypasses the need to deal with the genes one by one. It is often used subversively by bacterial viruses to take over the host polymerase and activate several sets of viral genes rapidly and sequentially (Figure 7-63).
Table 7-2
Sigma Factors of E. coli.

Figure 7-63
Interchangeable RNA polymerase subunits as a strategy to control gene expression in a bacterial virus. The bacterial virus SPO1, which infects the bacterium B. subtilis, uses the bacterial polymerase to transcribe its early genes immediately after the (more...)
In a sense, eucaryotes employ an analogous strategy through the use of three distinct RNA polymerases (I, II, and III) that share some of their subunits. Procaryotes, in contrast, use only one type of core RNA polymerase molecule, but they modify it with different sigma subunits.
Gene Switches Have Gradually Evolved
We have seen that the control regions of eucaryotic genes are often spread out over long stretches of DNA, whereas those of procaryotic genes are typically closely packed around the start point of transcription. Several bacterial gene regulatory proteins, however, recognize DNA sequences that are located many nucleotide pairs away from the promoter, as we saw in Figure 7-40. This case provided one of the first examples of DNA looping in gene regulation and greatly influenced later studies of eucaryotic gene regulatory proteins.
It seems likely that the close-packed arrangement of bacterial genetic switches developed from more extended forms of switches in response to the evolutionary pressure on bacteria to maintain a small genome size. This compression comes at a price, however, as it restricts the complexity and adaptability of the control device. The extended form of eucaryotic control regions, in contrast, with discrete regulatory modules separated by long stretches of spacer DNA, would be expected to facilitate a reshuffling of the regulatory modules during evolution, both to create new regulatory circuits and to modify old ones. Unraveling the history of how gene control regions evolved presents a fascinating challenge, and many clues can be found in present-day DNA sequences. We shall take up this issue again at the end of this chapter.
Summary
The transcription of individual genes is switched on and off in cells by gene regulatory proteins. In procaryotes these proteins usually bind to specific DNA sequences close to the RNA polymerase start site and, depending on the nature of the regulatory protein and the precise location of its binding site relative to the start site, either activate or repress transcription of the gene. The flexibility of the DNA helix, however, also allows proteins bound at distant sites to affect the RNA polymerase at the promoter by the looping out of the intervening DNA. Such action at a distance is extremely common in eucaryotic cells, where gene regulatory proteins bound to sequences thousands of nucleotide pairs from the promoter generally control gene expression. Eucaryotic activators and repressors act by a wide variety of mechanisms—generally causing the local modification of chromatin structure, the assembly of the general transcription factors at the promoter, and the recruitment of RNA polymerase.
Whereas the transcription of a typical procaryotic gene is controlled by only one or two gene regulatory proteins, the regulation of higher eucaryotic genes is much more complex, commensurate with the larger genome size and the large variety of cell types that are formed. The control region of the Drosophila eve gene, for example, encompasses 20,000 nucleotide pairs of DNA and has binding sites for over 20 gene regulatory proteins. Some of these proteins are transcriptional activators, whereas others are transcriptional repressors. These proteins bind to regulatory sequences organized in a series of regulatory modules strung together along the DNA, and together they cause the correct spatial and temporal pattern of gene expression. Eucaryotic genes and their control regions are often surrounded by insulators, DNA sequences recognized by proteins that prevent cross-talk between independently regulated genes.
- The Tryptophan Repressor Is a Simple Switch That Turns Genes On and Off in Bacteria
- Transcriptional Activators Turn Genes On
- A Transcriptional Activator and a Transcriptional Repressor Control the lac Operon
- Regulation of Transcription in Eucaryotic Cells Is Complex
- Eucaryotic Gene Regulatory Proteins Control Gene Expression from a Distance
- A Eucaryotic Gene Control Region Consists of a Promoter Plus Regulatory DNA Sequences
- Eucaryotic Gene Activator Proteins Promote the Assembly of RNA Polymerase and the General Transcription Factors at the Startpoint of Transcription
- Eucaryotic Gene Activator Proteins Modify Local Chromatin Structure
- Gene Activator Proteins Work Synergistically
- Eucaryotic Gene Repressor Proteins Can Inhibit Transcription in Various Ways
- Eucaryotic Gene Regulatory Proteins Often Assemble into Complexes on DNA
- Complex Genetic Switches That Regulate Drosophila Development Are Built Up from Smaller Modules
- The Drosophila eve Gene Is Regulated by Combinatorial Controls
- Complex Mammalian Gene Control Regions Are Also Constructed from Simple Regulatory Modules
- Insulators Are DNA Sequences That Prevent Eucaryotic Gene Regulatory Proteins from Influencing Distant Genes
- Bacteria Use Interchangeable RNA Polymerase Subunits to Help Regulate Gene Transcription
- Gene Switches Have Gradually Evolved
- Summary
- How Genetic Switches Work - Molecular Biology of the CellHow Genetic Switches Work - Molecular Biology of the Cell
Your browsing activity is empty.
Activity recording is turned off.
See more...